Module 1, midterm simulation¶
Download the data¶
Download [sciprog-qcb-2025-11-04-FIRSTNAME-LASTNAME-ID.zip] from Examina and extract it on your desktop. Folder content should be like this:
|- sciprog-qcb-2025-11-04-FIRSTNAME-LASTNAME-ID
|- QCB_midTerm_simulation.pdf
|- exercise1.py
|- exercise2.py
|- data_reduced.agp
|- small_seq.fasta
|- small.agp
Rename
sciprog-qcb-2025-11-04-FIRSTNAME-LASTNAME-IDfolder: put your name, lastname an id number, likesciprog-qcb-2025-11-04-john-doe-432432
From now on, you will be editing the files in that folder. At the end of the exam, that is what will be evaluated.
Edit the files following the instructions.
Problem 1¶
Given a list of positive integers (possibly repeated and unsorted) in the range [1,N], write a function that finds the missing values and returns them as a list. Note that the function should not crash if the list is empty. A warning should also be printed in case the user by mistake had negative numbers in the list.
Ex.
S = [1,9,7, 7, 4, 3, 3, 3]
S1 = list(range(10))
print(find_missing(S))
print(find_missing(S1))
print(find_missing([]))
S2 = [1, -72, 4, -3, -3, 3,10]
M = find_missing(S2)
print(M)
should return:
[2, 5, 6, 8]
[]
Warning: list is empty. Returning None
None
Warning -72 is <0. Ignoring it.
Warning -3 is <0. Ignoring it.
Warning -3 is <0. Ignoring it.
[2, 5, 6, 7, 8, 9]
Problem 2¶
The .agp file data_reduced.agp is a compact representation on how a set of assembled contigs made it into the scaffolds. The first few lines are reported below:
ScaffID s_start s_end type contig c_start c_end c_strand
scaffold_1 1 120953 W scf7180000021845 1 120953 -
scaffold_1 120954 121453 N 500 scaffold yes na
scaffold_1 121454 1026498 W scf7180000018491_2 1 905045 +
scaffold_1 1026499 1026998 N 500 scaffold yes na
In particular, the first row states that scaffold_1 from position 1 to 120953 has been built using the sequence of the contig scf7180000021845 from position 1 to 120953 in reverse strand (-) which means that the sequence has to be reverse-complemented. The second row states that in scaffold_1 positions 120954 to 121453 are a gap made of 500 N (note the 4th column is N rather W that stands for whole genome sequence).
Let’s suppose to have three sequences \(s1="ATAATA"\), \(s2="AAA"\) and \(s3="CCAAA"\), the following agp-formatted entries can be used to create a sequence my_scaff:
my_scaff 1 6 W s1 1 6 +
my_scaff 7 9 N 3 scaffold yes na
my_scaff 10 12 W s2 1 3 -
my_scaff 13 15 N 3 scaffold yes na
my_scaff 16 17 W s3 1 2 +
this would represent a fasta-formatted sequence:
>my_scaff
ATAATANNNTTTNNNCC
where basically the sequence is composed by s1 as it is, followed by three N, followed by the reverse complement of s2, three N and the first two characters of s3.
The file small_seq.fasta stores sequence information in .fasta format. A mock entry is the following:
>Chr01
AGGCCTAGGTCTTCCAGAGTCGCTTTTTCCAGCTCCAGACCGATCTCTTCAG
AGGCCAATCGCCAGTTTACCACATACACCCAGACCGATCTCTTCAG
where the first line is the identifier of the read and starts with a “>”. The sequence follows the line with the identifier and can be on multiple lines.
Implement the following python functions:
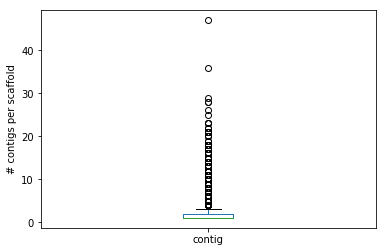
computeStats(filename, show_output = True): gets thefilenameof a .agp file as explained above, stores its content in a suitable data structure of your choice (hint: pandas might help here). Ifshow_outputisFalsethe function only returns the data structure. Otherwise, it counts (and prints) the total number of entries, the total number of scaffolds (hint: you can use DataFrame[column].unique()), total number of contigs (and their total size note that you might have to convert the c_start and c_end column to int with .astype(int)) and total number of gaps (and their total size). The function should also produce a box plot of the number of contigs per scaffold.
Note: The function should return the data structure containing all the data.
Calling:
fn = "data_reduced.agp"
scaffDF = computeStats(fn)
should give:
The file contains 7898 entries
... 1958 scaffolds
... 4928 contigs (tot. size: 873,456,804 bases)
... and 2970 gaps (tot. size: 1,485,000 bases)
printSequence(scaffInfo, scafID, sequenceFile): gets thescaffInfodata structure created by computeStats, a scaffold identifierscaffIDand the filename of a fasta formatted filesequenceFileand ifscafIDis present inscaffInfoit prints a fasta-formatted string reporting the sequence of the scaffold built as discussed above.
Hint: you can use biophtyon to read the fasta file.
Calling:
scaffDF = computeStats("small.agp", show_output = False)
printSequence(scaffDF,"my_scaff","small_seq.fasta")
print("")
printSequence(scaffDF,"my_scaff2","small_seq.fasta")
print("")
printSequence(scaffDF,"my_other_scaff","small_seq.fasta")
print("")
printSequence(scaffDF,"scaffold3","small_seq.fasta")
should give:
>my_scaff
ATAATANNNTTTNNNCC
>my_scaff2
TATTTTTATATGTATGTAATNNNNNNNNNNTTTATATATA
Warning: scaffold my_other_scaff not present
>scaffold3
NNNNNNNNNNNNNNNNNNNNCCCCGGAGGTACCTCCGGGGCCCCGGAGGT
Show/Hide a possible solution